SIUe - Computational Statistics (STAT 575) - Problem Set 4
Problem 1
Use Metropolis Hasting algorithm to generate
Part (a)
Implement your accept-reject algorithm and Metropolis-hastings algorithms to get
a sample of
Acceptance-rejection sampler
We implement the density and sampler functions, respectively
Suppose
Suppose
We may thus use acceptance-rejection sampling to sample
Note that we may use their respective kernels instead,
since the ratio of the kernels is proportional to
We implement the acceptance-rejection algorithm with the following code:
# The density function for random variates in the family
# GAM(shape=alpha,rate=1)
dgamma1 <- function(x, alpha) {
dgamma(x, shape = alpha, rate = 1)
}
# An acceptance-rejection sampling procedure for random variates in the family
# GAM(shape=alpha,rate=1)
accept.reject.rgamma1 <- function(n, alpha) {
a <- floor(alpha)
rate <- a/alpha
q <- (alpha/exp(1))^(a - alpha)
ys <- vector(length = n)
for (i in 1:n) {
repeat {
y <- sum(rexp(a, rate)) # draw candidate
if (runif(1) <= q * y^(alpha - a) * exp(y * a/alpha - y)) {
ys[i] <- y
break
}
}
}
ys
}We sample from
alpha <- 2.5
m <- 10000
accept.reject.samp <- accept.reject.rgamma1(m, alpha)Metropolis-Hastings algorithm
Here is our implementation of the Metropolis-Hastings algorithm:
# A sampling procedure for random variates in the family
# GAM(shape=alpha,rate=1) using Metropolis-Hastings algorithm
metro.hast.rgamma1 <- function(n, alpha, burn = 0) {
a <- floor(alpha)
rate <- a/alpha
# density for random variates in the family GAM(shape=alpha,rate=1)
f <- function(x) {
dgamma(x, shape = alpha, rate = 1)
}
g <- function(x) {
dgamma(x, shape = a, rate = rate)
}
m <- n + burn
ys <- vector(length = m)
ys[1] <- sum(rexp(a, rate))
for (i in 2:m) {
v <- sum(rexp(a, rate)) # draw from g
u <- ys[i - 1]
R <- f(v) * g(u)/(f(u) * g(v))
if (runif(1) <= R) {
ys[i] <- v
} else {
ys[i] <- u
}
}
ys[(burn + 1):m]
}We sample from the Metro-Hastings algorithm with the following code:
metro.hast.samp <- metro.hast.rgamma1(m, alpha)Part (b)
Check on mixing and convergence using plots. Run multiple chain and compute the Gelman-Rubin statistics. You may pick any reasonable burn-in.
We plot the histograms with:
par(mfrow = c(1, 2))
hist(accept.reject.samp, freq = F, breaks = 50, main = "acceptance-rejection")
lines(seq(0.01, 15, by = 0.01), dgamma1(seq(0.01, 15, by = 0.01), alpha), col = "blue",
lwd = 2)
hist(metro.hast.samp, freq = F, breaks = 50, main = "metropolis-hastings")
lines(seq(0.01, 15, by = 0.01), dgamma1(seq(0.01, 15, by = 0.01), alpha), col = "blue",
lwd = 2)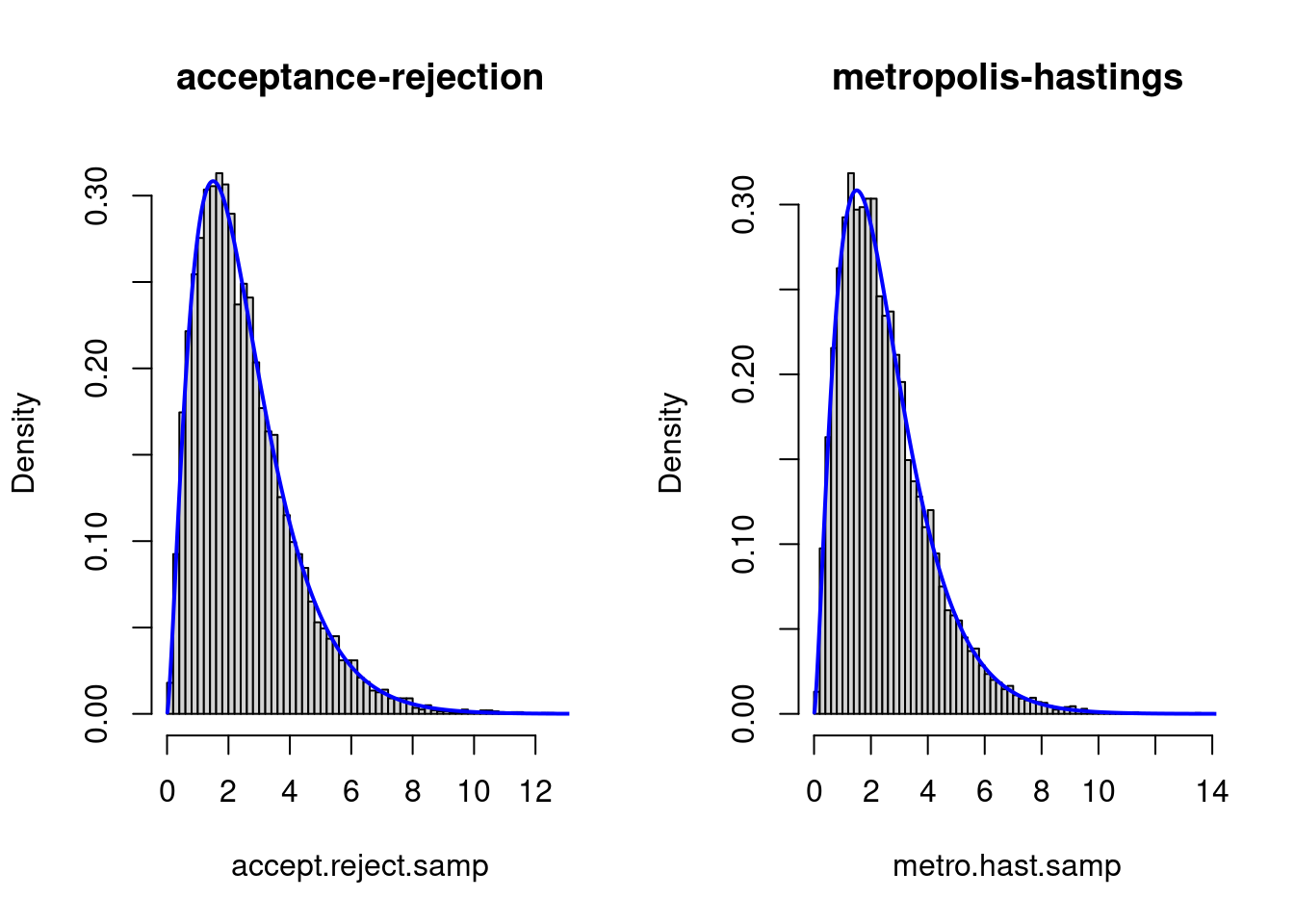
Both samplers seem to be compatible with the density.
We plot the of the Metropolis-Hastings and acceptance-rejection samplers with:
par(mfrow = c(1, 2))
plot(metro.hast.samp, pch = "·", xlab = "t", ylab = "Y", main = "metropolis-hastings")
plot(accept.reject.samp, pch = "·", xlab = "t", ylab = "Y", main = "acceptance-rejection")
Both of these look good, as neither remain at or near the same value for many iterations. They also both quickly move away from their initial values.
We plot the ACFs with:
par(mfrow = c(1, 2))
acf(metro.hast.samp)
acf(accept.reject.samp)
Both samplers seem to have very little autocorrelation. If an uncorrelated sample is extremely important, to be safe, taking every other sample point would probably be sufficient.
We implement the Gelman-Rubin statistic with:
# samps: should be an L x J matrix, where L is the length of the samples and J
# is the number of samples (independent chains).
gelman.rubin <- function(samps) {
L <- nrow(samps)
J <- ncol(samps)
x.bar <- apply(samps, 2, mean)
B <- var(x.bar) * L
W <- mean(apply(samps, 2, var))
((L - 1)/L * W + B/L)/W
}Next, we compute Gelman-Rubin statistics on the computed independence chains.
chains <- 1000
samps <- matrix(nrow = m, ncol = chains)
for (i in 1:chains) {
samps[, i] <- metro.hast.rgamma1(m, alpha, burn = 1000)
}
gelman.rubin.stat <- gelman.rubin(samps)
print(gelman.rubin.stat)## [1] 1.000009We see that the Gelman-Rubin statistic is given by
We adopt the rule of thumb that if
Part (c)
Estimate
Theoretically,
We estimate
tab <- matrix(nrow = 2, ncol = 1)
rownames(tab) <- c("acceptance-rejection", "metropolis-hastings")
colnames(tab) <- c("mean")
tab[1] <- c(mean(accept.reject.samp^2))
tab[2] <- c(mean(metro.hast.samp^2))
knitr::kable(data.frame(tab))| mean | |
|---|---|
| acceptance-rejection | 8.743824 |
| metropolis-hastings | 8.703302 |
Both are quite close to the true value of
Problem 2 (Problem 7.1)
Rework the textbook example. Consider the mixture normal
Part (a)
Simulate
We implement the density and sampler for the mixture distribution with:
dmix <- function(x, delta) {
delta * dnorm(x, 7, 0.5) + (1 - delta) * dnorm(x, 10, 0.5)
}
rmix <- function(n, delta) {
xs <- vector(length = n)
for (i in 1:n) {
xs[i] <- ifelse(runif(1) < delta, rnorm(1, 7, 0.5), rnorm(1, 10, 0.5))
}
xs
}We generate a sample and plot its histogram with:
n <- 200
delta <- 0.7
data <- rmix(n, delta)
hist(data, freq = F)
Part (b)
Now assume
lmix <- Vectorize(function(delta, xs) {
if (delta < 0 || delta > 1) {
return(0)
}
p <- 1
for (x in xs) {
p <- p * dmix(x, delta)
}
p
}, "delta")
logmix <- Vectorize(function(delta, xs) {
if (delta < 0 || delta > 1) {
return(-Inf)
}
logp <- 0
for (x in xs) {
logp <- logp + log(dmix(x, delta))
}
logp
}, "delta")A sample
In the independence chain MCMC, we may use the prior as the proposal density,
Numerical imprecision
Suppose we have a data type
In the likelihood function,
Suppose
With the above in mind, we replace the likelihood function with the log-likelihood function to significantly increase the space of samples we can work with.
delta.estimator.ic <- function(n, data, delta0 = runif(1), burn = 0) {
m <- n + burn
deltas <- vector(length = m)
deltas[1] <- delta0
for (i in 2:m) {
delta <- runif(1) # draw candidate from prior
delta.old <- deltas[i - 1]
log.R <- logmix(delta, data) - logmix(delta.old, data)
if (log(runif(1)) <= log.R) {
deltas[i] <- delta
} else {
deltas[i] <- delta.old
}
}
deltas[(burn + 1):m]
}Part (c)
Implement a random walk chain with
We observe that
delta.estimator.rw <- function(n, data, delta0 = runif(1), burn = 0) {
m <- n + burn
deltas <- vector(length = m)
deltas[1] <- delta0
for (i in 2:m) {
delta.old <- deltas[i - 1]
delta <- delta.old + runif(1, -1, 1)
log.R <- logmix(delta, data) - logmix(delta.old, data)
if (log(runif(1)) <= log.R) {
deltas[i] <- delta
} else {
deltas[i] <- delta.old
}
}
deltas[(burn + 1):m]
}Part (d)
Reparameterize the problem letting
logit <- function(delta) {
log(delta/(1 - delta))
}
logit.inv <- function(u) {
exp(u)/(1 + exp(u))
}
logit.inv.J <- function(u) {
exp(u)/(1 + exp(u))^2
}
delta.estimator.u.rw <- function(n, data, delta0 = runif(1), burn = 0) {
m <- n + burn
u <- vector(length = m)
u[1] <- logit(delta0)
for (i in 2:m) {
u.old <- u[i - 1]
u.star <- u.old + runif(1, -1, 1)
R <- lmix(logit.inv(u.star), data) * logit.inv.J(u.star)/(lmix(logit.inv(u.old),
data) * logit.inv.J(u.old))
if (runif(1) <= R) {
u[i] <- u.star
} else {
u[i] <- u.old
}
}
logit.inv(u[(burn + 1):m])
}Part (e)
Compare the estimates and convergence behavior of three algorithms.
We do not do a burn-in, since we are interested in seeing how quickly
the three methods converge. We only plot chains of length
We generate the data sets with:
chain <- 1000
burn <- 0
deltas.ic <- delta.estimator.ic(chain, data, burn = burn)
deltas.rw <- delta.estimator.rw(chain, data, burn = burn)
deltas.u.rw <- delta.estimator.u.rw(chain, data, burn = burn)
tab <- matrix(nrow = 3, ncol = 1)
rownames(tab) <- c("independence chain", "random walk", "reparameterized random walk")
colnames(tab) <- c("mu")
tab[1, ] <- mean(deltas.ic)
tab[2, ] <- mean(deltas.rw)
tab[3, ] <- mean(deltas.u.rw)
knitr::kable(data.frame(tab))| mu | |
|---|---|
| independence chain | 0.6887205 |
| random walk | 0.6959496 |
| reparameterized random walk | 0.6950262 |
As the table of estimations shows, all three methods provide a good estimate
of
We plot the histograms with:
par(mfrow = c(1, 3))
hist(deltas.ic, freq = F, breaks = 50, main = "independence chain")
hist(deltas.rw, freq = F, breaks = 50, main = "random walk")
hist(deltas.u.rw, freq = F, breaks = 50, main = "reparameterized random walk")
The reparameterized random walk metropolis has a histogram that is most
compatible with normality, i.e., characteristic bell curve with a mode at
We plot the sample paths with:
par(mfrow = c(1, 3))
plot(deltas.ic, pch = "·", type = "l", xlab = "t", ylab = "delta", main = "independence chain")
plot(deltas.rw, pch = "·", type = "l", xlab = "t", ylab = "delta", main = "random walk")
plot(deltas.u.rw, pch = "·", type = "l", xlab = "t", ylab = "delta", main = "reparameterized random walk")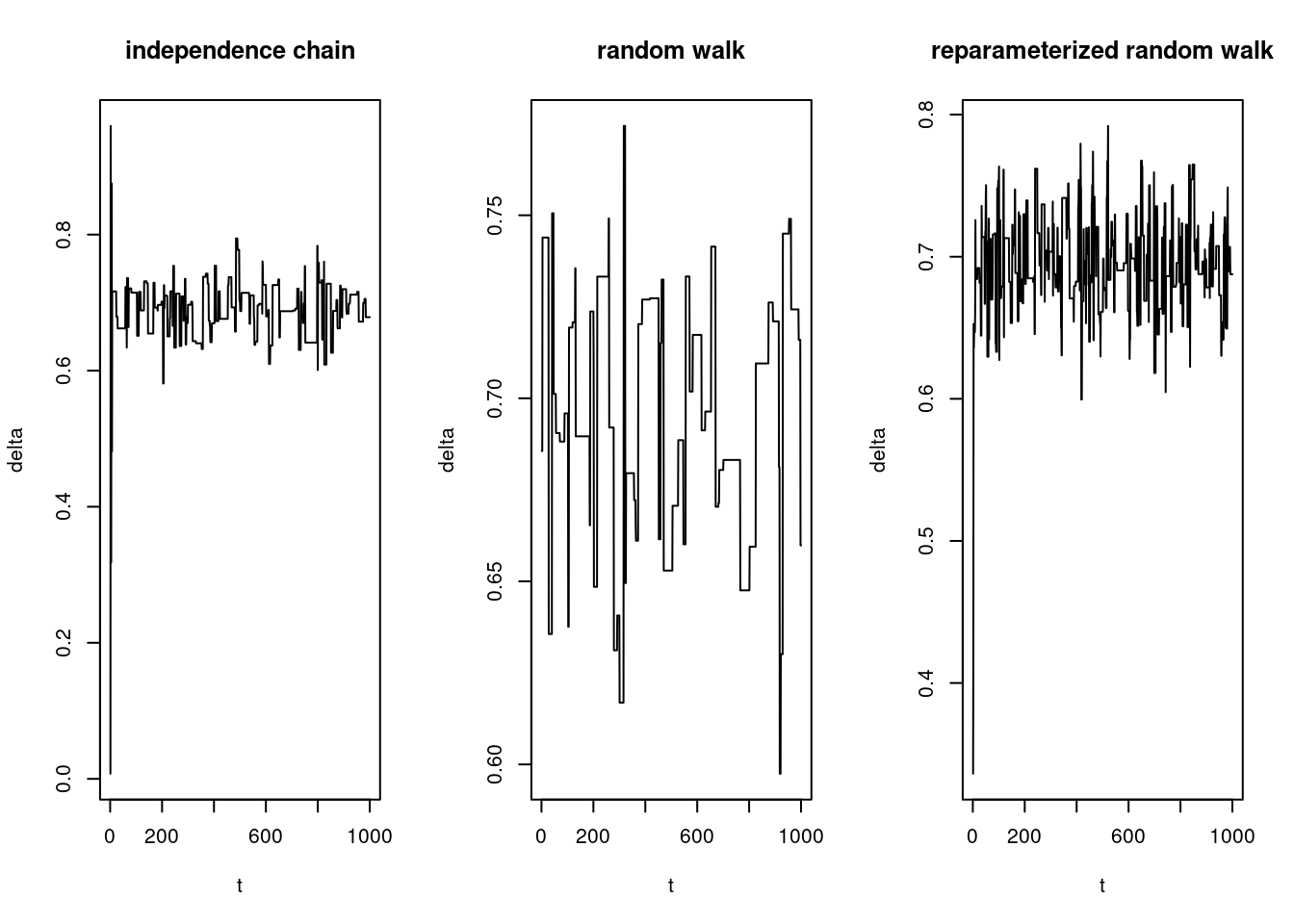
We see that the random walk demonstrates relatively poor mixing. It has a high rejection rate (stays at the same level for long periods of time), causing it to explore the support of the likelihood slowly.
The sample path of the indepedence chain also can be said to demonstrate poor mixing.
The reparameterized random walk exihibits good mixing, vigorously jiggling around the true value.
We plot the ACFs with:
par(mfrow = c(1, 3))
acf(deltas.ic, main = "independence chain")
acf(deltas.rw, main = "random walk")
acf(deltas.u.rw, main = "reparameterized random walk")
They all decay quickly, but in order of increasing autocorrelation: the reparameterized random walk, the independence chain, and the random walk.
In particular, the reparameterized random walk shows autocorrelation that decays quite rapidly with respect to lag time.
Problem 3
Consider an i.i.d. sample
Part (a)
Write out the posterior distribution of
Note that the posterior distribution is given by
The likelihood function of
Part (b)
Show the posterior conditional distribution of
Distribution of
The conditional distribution of
Now, we wish to show that this is the kernel of the given normal distribution.
We do this by discarding any factors that are not a function of
Completing the square, we obtain
Thus, we see that this is the kernel of a normal density with mean
Distribution of
The conditional distribution of
We seek to match it to the kernel of the gamma density for
We may rewrite
We note that
Part (c)
First, generate some ``observed’’ sample data using
We generate the sample with:
x <- rnorm(200, mean = 5, sd = 2)We implement the Gibbs sampling with the function:
mu.tau.gibbs <- function(n, x, burn = 1000, theta0 = NULL, p = 1e-04, m = 0, a = 1e-04,
b = 1e-04) {
x.mu <- mean(x)
x.s2 <- var(x)
x.n <- length(x)
rmu <- function(tau) {
mean <- (x.n * tau * x.mu + p * m)/(x.n * tau + p)
var <- 1/(x.n * tau + p)
rnorm(1, mean = mean, sd = sqrt(var))
}
rtau <- function(mu) {
rgamma(1, shape = a + x.n/2, rate = b + x.n * (x.s2 + (mu - x.mu)^2)/2)
}
prior <- function() {
c(rnorm(1, m, 1/p), rgamma(1, a, b))
}
N <- n + burn
thetas <- matrix(nrow = N, ncol = 2)
if (is.null(theta0)) {
thetas[1, ] <- prior()
} else {
thetas[1, ] <- theta0
}
for (i in 1:(N - 1)) {
tau.new <- rtau(thetas[i, 1])
mu.new <- rmu(tau.new)
thetas[i + 1, ] <- c(mu.new, tau.new)
}
thetas <- thetas[(burn + 1):N, ]
mu.est <- mean(thetas[, 1])
tau.est <- mean(thetas[, 2])
sigma.est <- sqrt(1/tau.est)
list(theta.dist = thetas, mu.est = mu.est, tau.est = tau.est, sigma.est = sigma.est)
}We use the Gibbs sampler to estimate
# set up hyper-parameters
a <- 1e-04
b <- 1e-04
m <- 0
p <- 1e-04
res <- mu.tau.gibbs(1e+05, x, burn = 10000, p = p, a = a, m = m, b = b)
mu.est <- round(as.numeric(res$mu.est), digits = 4)
tau.est <- round(as.numeric(res$tau.est), digits = 4)
var.est <- round(1/tau.est, digits = 4)
c(mu.est, tau.est)## [1] 4.8905 0.2633We estimate
Additional analysis
Out of curiosity, we decided to plot the marginals of the sample superimposed with their respective conditional densities with:
x.mu <- mean(x)
x.n <- length(x)
x.s2 <- var(x)
mu.dist <- res$theta.dist[, 1]
tau.dist <- res$theta.dist[, 2]
hist(mu.dist, freq = F, breaks = 50, main = "marginal of mu", xlab = "mu")
lines(seq(4, 6, by = 0.01), dnorm(seq(4, 6, by = 0.01), mean = (x.n * tau.est * x.mu)/(x.n *
tau.est + p), sd = sqrt(1/(x.n * tau.est + p))))
hist(tau.dist, freq = F, breaks = 50, main = "marginal of tau", xlab = "tau")
lines(seq(0.15, 0.4, by = 0.01), dgamma(seq(0.15, 0.4, by = 0.01), shape = a + x.n/2,
rate = b + x.n * (x.s2 + (mu.est - x.mu)^2)/2))