SIUe - Computational Statistics (STAT 575) - Problem Set 1
Problem 1
Write your own code and find solution to the equation \(x^3 + x - 4 = 0\) using Newton’s method and the secant method. Compare the number of iterations needed for different starting values for the two methods.
Solution: Newton’s method
If we have some function \(f : \mathbb{R} \mapsto \mathbb{R}\) and we wish to find a root of \(f\), i.e., an \(x\) such that \(f(x) = 0\), we may use Newton’s method.
We take an initial guess of the root as \(x_0\) and try to refine it with a linear approximation of \(f\) given by \[ L(f | x_0) = \lambda x.f(x_0) + f'(x_0)(x-x_0). \]
Now, we may approximate a root of \(f\) with a root of \(L(f|x_0)\), \[ L(f|x_0)(x) = 0, \] which may be rewritten as \[ f(x_0) + f'(x_0)(x-x_0) = 0. \] Solving for a root \(x\) of \(L(f|x_0)\) we get the result \[ x = x_0 - \frac{f(x_0)}{f'(x_0)}. \]
Hoping that \(x\) results in a better approximation of the root of \(f\) than \(x_0\), we approximate \(f\) with \(L(f|x)\) and repeat the process.
Generalizing this result, we obtain the iterative procedure \[ x_{i+1} = x_i - \frac{f(x_i)}{f'(x_i)}. \] We continue this process until we obtain some stopping condition, e.g., \(|x_{i+1} - x_i| < \epsilon\).
Letting \(f(x) = x^3 + x - 4\) and \(f'(x) = 3x^2 + 1\) and substituting into the above result, we get the result \[ x_{i+1} = x_i - \frac{x_i^3+x_i-4}{3x_i^2+1}. \]
We implement a general procedure for Newton’s method:
newton_method <- function(f,dfdx,x0,eps,debug=T)
{
n <- 0
repeat
{
x1 <- x0 - f(x0) / dfdx(x0)
n <- n + 1
if (debug==T) { cat("iteration=",n," x=",x1,"\n") }
if(abs(x1 - x0) < eps)
{
break
}
x0 <- x1
}
list(root=x0,iter=n)
}We take an initial guess of \(x_0 = 1\) and \(\epsilon = 1 \times 10^{-6}\) and run the following R code to solve for a root of \(f\) using Newton’s method:
f <- function(x) { x^3 + x - 4 }
dfdx <- function(x) { 3*x^2 + 1 }
eps <- 1e-6
x0 <- 1
result <- newton_method(f,dfdx,x0,eps)## iteration= 1 x= 1.5
## iteration= 2 x= 1.387097
## iteration= 3 x= 1.378839
## iteration= 4 x= 1.378797
## iteration= 5 x= 1.378797We obtain \(x \approx 1.3787967\) after \(5\) iterations. When we plug that approximate root into \(f\) we obtain the result \(f(1.3787967) = 7.3825959\times 10^{-9}\), which is approximately zero.
Solution: Secant method
In Newton’s method, we linearize \(f\) using the derivative of \(f\). If, instead, we use the secant of \(f\) with respect to two inputs \(x_i\) and \(x_{i+1}\), as given by \[ \frac{f(x_{i+1}) - f(x_i)}{x_{i+1}-x_i}, \] we get the iterative procedure \[ x_{i+2} = x_{i+1} - f(x_{i+1})\frac{x_{i+1}-x_i}{f(x_{i+1}) - f(x_i)}, \] which requires two initial values \(x_0\) and \(x_1\).
We define the secant method as a function given by:
secant_method <- function(f,x0,x1,eps,debug=T)
{
n <- 0
repeat
{
x2 <- x1 - f(x1) * (x1 - x0) / (f(x1) - f(x0))
n <- n + 1
if (debug==T) { cat("iteration=",n," x=",x2,"\n") }
if(abs(x2-x1) < eps)
{
break
}
x0 <- x1
x1 <- x2
}
list(root=x1,iter=n)
}We let \(x_0 = 0\), \(x_1 = 1\), and keep everything else the same and run the secant method with the following R code:
x0 <- 0
x1 <- 1
result <- secant_method(f,x0,x1,eps,F)We obtain \(x \approx 1.3787965\) after \(7\) iterations. When we plug that approximate root into \(f\) we obtain the result \(f(1.3787965) = -1.268648\times 10^{-6}\), which is approximately zero.
Note that this is \(2\) more iterations than Newton’s method.
Comparison of Newton’s method versus secant method
We perform \(10000\) trials to get a better view of how the two methods, Newton and secant, compare over many different initial guesses.
We generate the data with:
n <- 10000
from <- 0
to <- 4
by <- (to-from)/n
newt_sols <- vector(length=n)
sec_sols <- vector(length=n)
i <- 1
for (x0 in seq(from=from, to=to, by=by))
{
newt_sols[i] <- newton_method(f,dfdx,x0,eps,F)$iter
sec_sols[i] <- secant_method(f,x0,x0+1,eps,F)$iter
i <- i + 1
}We summarize the results and report them with:
cat("mean iterations\n",
"newton => ", mean(newt_sols), "\n",
"secant => ", mean(sec_sols), "\n")## mean iterations
## newton => 5.819618
## secant => 7.231077We see that Newton’s method, on average, requires \(1.4114589\) fewer iterations before the stopping condition is satisfied.
Problem 2
Poisson regression. The Ache hunting data set has \(n = 47\) observations recording is the number of monkeys killed over a period of days with each hunter along with hunter’s age. It is of interest to estimate and quantify the monkey kill rate as a function of hunter’s age. Hunting prowess confers elevated status among the group, so a natural question is whether hunting ability improves with age, and at which age hunting ability is best.
Hand-code Newton-Raphson in R to fit the Poisson regression model \[ \mathit{monkeys}_i \sim \operatorname{Pois}\left(\exp(\log \mathit{days}_i + \theta_1 + \theta_2 \mathit{age}_i + \theta_3 \mathit{age}_i^2)\right). \]
Feel free to use jacobian and hessian in the numDeriv R package. You may need a sets of crude starting values. I run a linear regression for the “empirical log-rates” and get starting values \((5.99, 0.167, 0.001)\). Feel free to use those. Compare your result with glm() function in R using
glm(monkeys~age+I(age^2), family="poisson", offset=log(days), data=d)Solution
We are given the following data:
d <- read.table("ache.txt", header=T)
n <- length(d$age)
X <- cbind(rep(1,n), d$age, d$age^2)
loglike <- function(theta)
{
sum(dpois(d$monkeys,exp(log(d$days)+X%*%theta),log=T))
}We generalize the univariate Newton’s method in Problem 1 to the multivariate case. We implement the multivariate Newton-Raphson method with numerical hessian and jacobian with the following R code:
library(numDeriv)
newton_raphson_method <- function(x0,f,eps)
{
n <- 0
x1 <- x0
repeat
{
x1 <- x0 - solve(hessian(f,x0))%*%t(jacobian(f,x0))
n <- n + 1
if (n %% 7 == 0) { cat("iteration=",n," theta=",x1,"\n") }
if (max(abs(x1 - x0)) < eps)
{
break
}
x0 <- x1
}
list(root=x1,iter=n)
}We use the multivariate Newton-Raphson method to find the MLE of \(\theta\) in the poisson regression model:
eps <- 1e-6
theta0 <- c(5.99, 0.167, 0.001) # starting values
theta_mle <- newton_raphson_method(theta0,loglike,eps)$root## iteration= 7 theta= -1.01133 0.1670442 0.0009996348
## iteration= 14 theta= -7.590958 0.1532685 0.001112005
## iteration= 21 theta= 1.438482 -0.2696711 0.003827083
## iteration= 28 theta= -5.484246 0.1246477 -0.001203418The MLE of \(\theta\) is given by:
theta_mle## [,1]
## [1,] -5.484245904
## [2,] 0.124647667
## [3,] -0.001203418We compare the results with the builtin method:
glm(monkeys~age+I(age^2),family="poisson", offset=log(days),data=d)$coefficients## (Intercept) age I(age^2)
## -5.484245904 0.124647667 -0.001203418The hand-coded approach and the builtin approach obtain the same point estimate \(\hat\theta = (-5.4842, 0.1246, -0.0012)'\).
Problem 3
Logistic and Cauchy distributions are well-suited to the inverse transform method. For each of the following, generate \(10,000\) random variables using the inverse transform. Compare your program with the built-in R functions rlogis() and rcauchy(), respectively:
Solution: part (a)
Standard logistic distribution \[ F(x) = \frac{1}{1+e^{-x}} \]
Solve for \(x\) in \[\begin{align*} u &= F(x)\\ u &= \frac{1}{1+e^{-x}}\\ x &= \log(u/(1-u)). \end{align*}\]
n <- 10000
us <- runif(n)
d1 <- density(log(us/(1-us)))
d2 <- density(rlogis(n))
plot(d1,col="blue",main="comparison of density plots")
lines(d2,col="red")
legend(x="topright",legend=c("inverse method","built-in"),col=c("blue","red"),
pch=c("-","-"))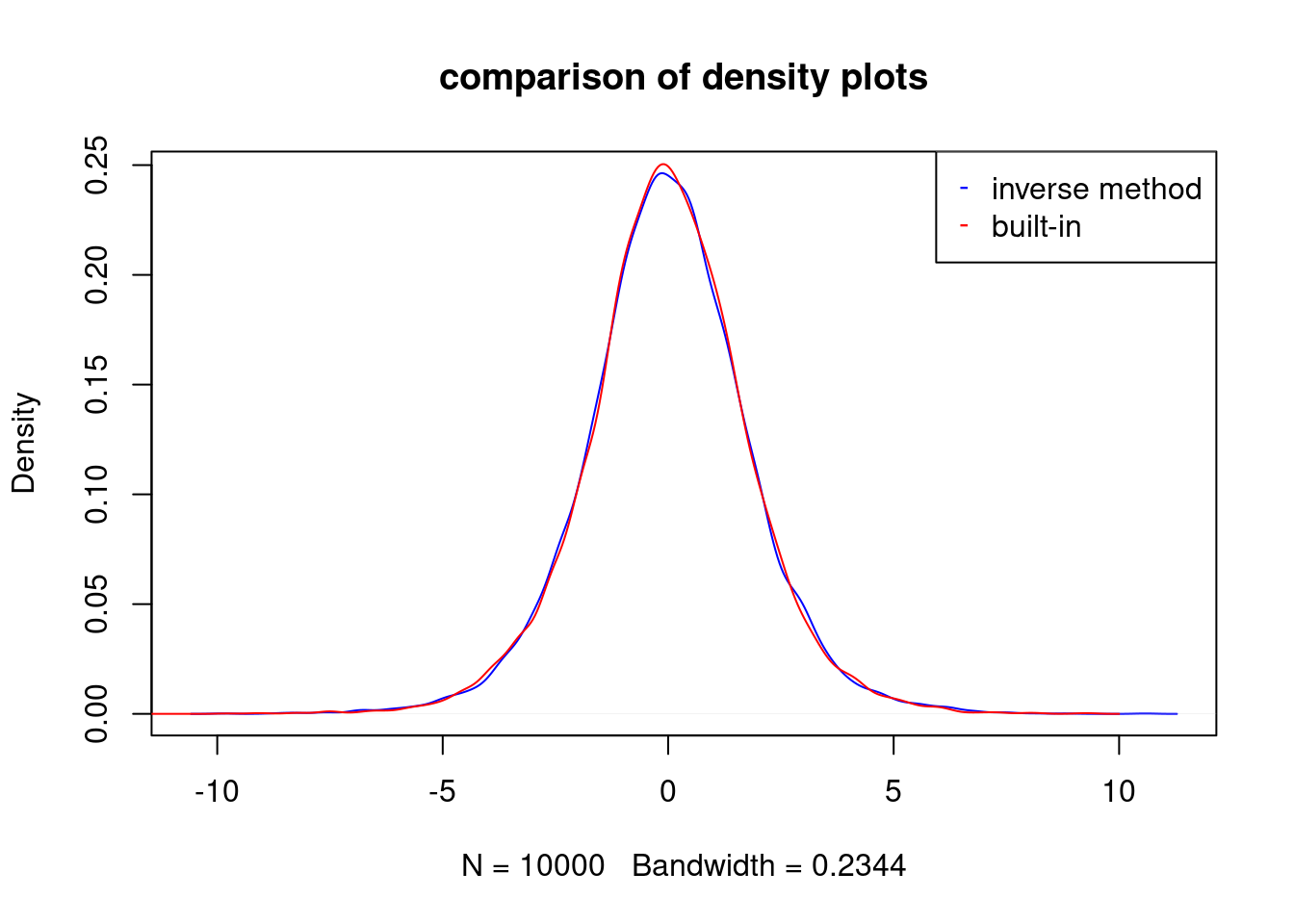
Solution: part (b)
Standard Cauchy distribution \[ F(x) = \frac{1}{2} + \frac{1}{\pi} \operatorname{arctan(x)} \]
Solve for \(x\) in \[\begin{align*} u &= F(x)\\ u &= \frac{1}{2} + \frac{1}{\pi} \operatorname{arctan(x)}\\ x &= \tan(\pi(u-1/2)). \end{align*}\]
n <- 1000
us <- runif(n)
d1 <- tan(pi*(us-0.5))
d2 <- rcauchy(n=n)
d1 <- d1[d1 > -20 & d1 < 20]
d2 <- d2[d2 > -20 & d2 < 20]
c1 <- rgb(0,0,255, max = 255, alpha = 50, names = "blue")
c2 <- rgb(255,0,0, max = 255, alpha = 50, names = "red")
par(mfrow=c(1,2))
hist(d1,col=c1,freq=F,breaks=50,main="inverse-transform method vs built-in")
hist(d2,col=c2,add=T,freq=F,breaks=50)
legend(x="topright",legend=c("inv","builtin"),col=c("blue","red"),pch=c("-","-"))
plot(density(d1), col="blue",main="density plot")
lines(density(d2), col="red")
legend(x="topright",legend=c("inv","builtin"),col=c("blue","red"),pch=c("-","-"))
Problem 4
Generating \(10,000\) random variables from \(\operatorname{Geometric}(p)\) distribution based off Bernoulli trials.
Solution
A random variable \(X \sim \operatorname{Geometric(p)}\) is given by the number of i.i.d. trials needed to have a success where success occurs with probability \(p\).
Thus, we may simulate this distribution with the following R code:
# simulate n realizes of geometric(p)
rgeo <- function(n,p)
{
outcomes <- vector(length=n)
for (i in 1:n)
{
trials <- 0
while (T)
{
trials <- trials + 1
if (rbinom(1,1,p) == 1)
{
break
}
}
outcomes[i] <- trials
}
outcomes
}When we use this function to draw a sample of \(n=10000\) geoemtrically distributed random variables with \(p=0.2\), we obtain:
p <- .2
n <- 10000
sample <- rgeo(n,p)
cat("the mean should be approximate 1/p =", 1/p, " and we obtain a mean of ", mean(sample))## the mean should be approximate 1/p = 5 and we obtain a mean of 5.0425Problem 5
Generate random values from a Standard Half Normal distribution with pdf, \[ f(x) = \frac{2}{\sqrt{2 \pi}} e^{-x^2/2} , x > 0. \]
For the candidate pdf, choose the exponential density with rate \(1\). Verify that your method works via a plot of the true density, and a histogram of the generated values.
Solution
We are given the density of the standard half-normal distribution, \[ \operatorname{dhalfnormal}(x) = \frac{2}{\sqrt{2 \pi}} e^{-x^2/2}, x > 0. \]
We model this density with the following R code:
# density for standard half-normal
dhalfnormal <- function(x) { 2/sqrt(2*pi)*exp(-x^2/2) }We sample from the exponential distribution \(\operatorname{EXP}(\lambda=1)\), with density \(g\) and thus we first find the \(c\) satisfying \[ c = \max \left\{ \frac{\operatorname{dhalfnormal}(x)}{\operatorname{dexp}(x|\lambda=1)} | x \in \mathbb{R} \right\}, \] which is found to be approximately \(c = 1.315489247\).
We implement the standard half-normal sampler, \(\operatorname{rhalfnormal}\), using the acceptance-rejection sampling technique with the following R code:
# accept-rejection sampling for standard half-normal
# using exp(rate=1)
rhalfnormal <- function(N)
{
c <- 1.315489247
xs <- vector(length=N)
k <- 1
while (T)
{
x <- rexp(n=1)
if (runif(n=1) < dhalfnormal(x)/(c*dexp(x)))
{
xs[k] <- x
k <- k + 1
if (k == N)
{
break
}
}
}
xs
}We simulate drawing \(n=100000\) samples from the standard half-normal distribution and plotting a histogram of the sample with its density overload in red on top of it with the following R code:
n <- 100000
sample <- rhalfnormal(n)
hist(sample,freq=F,breaks=50,main="standard half-normal")
curve(dhalfnormal(x),add=TRUE,col="red") 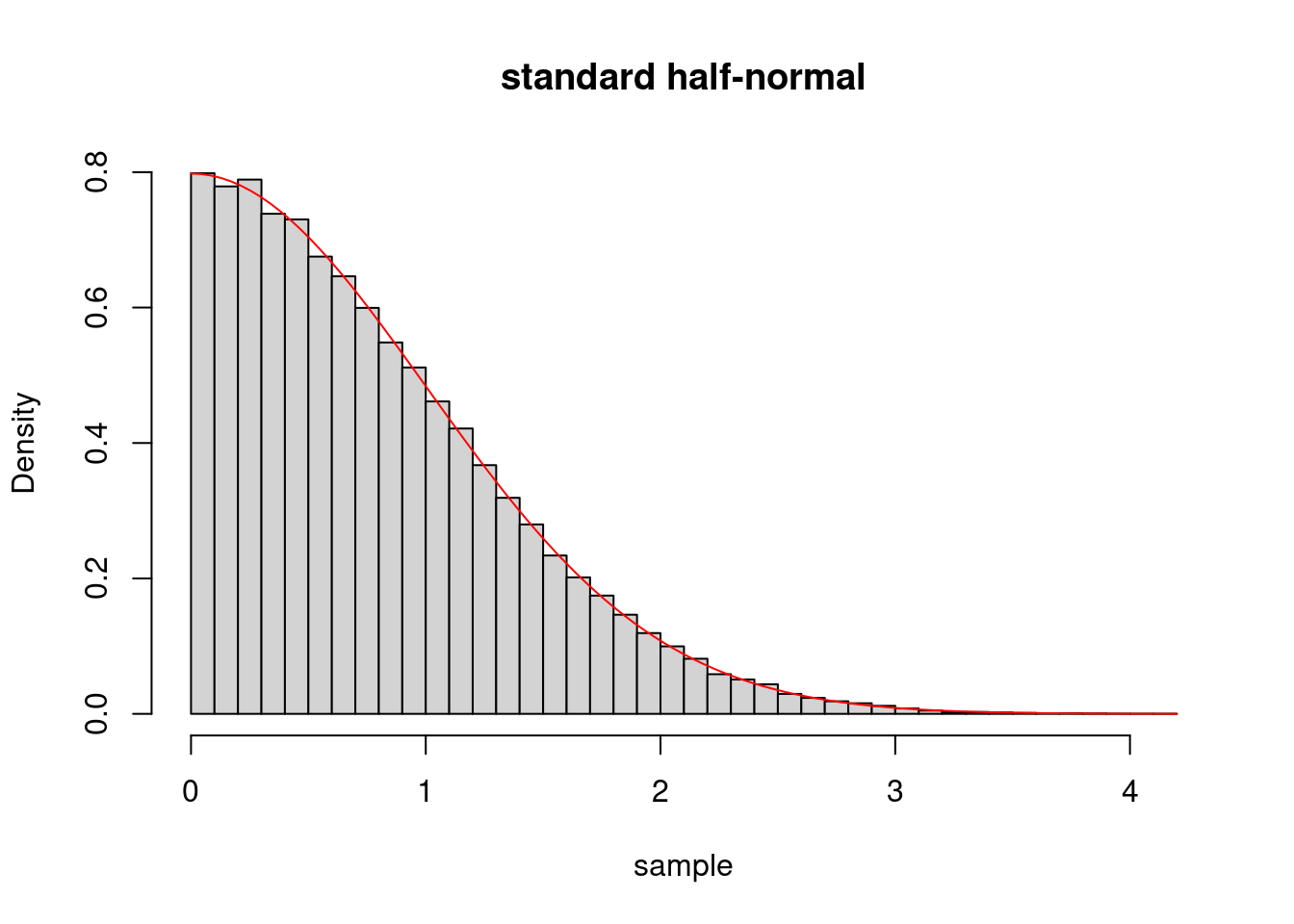
We see that the histogram is compatible with being drawn from the overload density.
Problem 6
Use accept-reject to sample from this bimodal density: \[ f(x) \propto 3 e^{-0.5(x+2)^2} + 7 e^{-0.5(x-2)^2} \] The normalizing constant is \(25.066\). For your proposal \(g(·)\), use a \(N(0, 2^2)\) distribution. Verify that your method works via a plot of the true normalized density, and a histogram of the generated values.
Solution
We are given the kernel of the bimodal distribution of interest, \[ \operatorname{ker-bimodal}(x) = 3 e^{-0.5(x+2)^2} + 7 e^{-0.5(x-2)^2}, \] with the normalizing constant \(C = 25.0663\) and thus the pdf for the bimodal is given by \[ \operatorname{dbimodal}(x) = \frac{\operatorname{ker}(x)}{C}. \]
We model these two functions with the following R code:
# density for biomodal density
kerbimodal <- function(x) { 3*exp(-0.5*(x+2)^2) + 7*exp(-0.5*(x-2)^2) }
kerbimodal.C <- 25.0663
dbimodal <- function(x) { kerbimodal(x) / kerbimodal.C }We sample from the normal distribution \(N(\mu=0,\sigma^2=2^2)\), with density \(g\) and thus we first find the \(c\) satisfying \[ c = \max \left\{ \frac{\operatorname{ker-bimodal}(x)}{g(x|\mu=0,\sigma^2=2^2)} | x \in \mathbb{R} \right\}, \] which is found to be approximately \(c = 68.35212\).
We implement the bimodal sampler, \(\operatorname{rbimodal}\), using the acceptance-rejection sampling technique with the following R code:
# accept-rejection sampling for bimodal distribution with density dbimodal
# using normal(0,2^2).
rbimodal <- function(N)
{
c <- 68.35212
xs <- vector(length=N)
k <- 1
while (T)
{
x <- rnorm(n=1,mean=0,sd=2)
if (runif(n=1) < kerbimodal(x)/(c*dnorm(x,mean=0,sd=2)))
{
xs[k] <- x
if (k == N)
{
break
}
k <- k + 1
}
}
xs
}We simulate drawing \(n=100000\) samples from the bimodal distribution and plotting a histogram of the sample with its density overload in red on top of it with the following R code:
n <- 100000
sample <- rbimodal(n)
hist(sample,freq=F,breaks=50,main="bimodal")
curve(dbimodal(x),add=TRUE,col="red") 
We see that the histogram is compatible with being drawn from the overload density.